

|
Prediction of microRNA target sites and more |
|
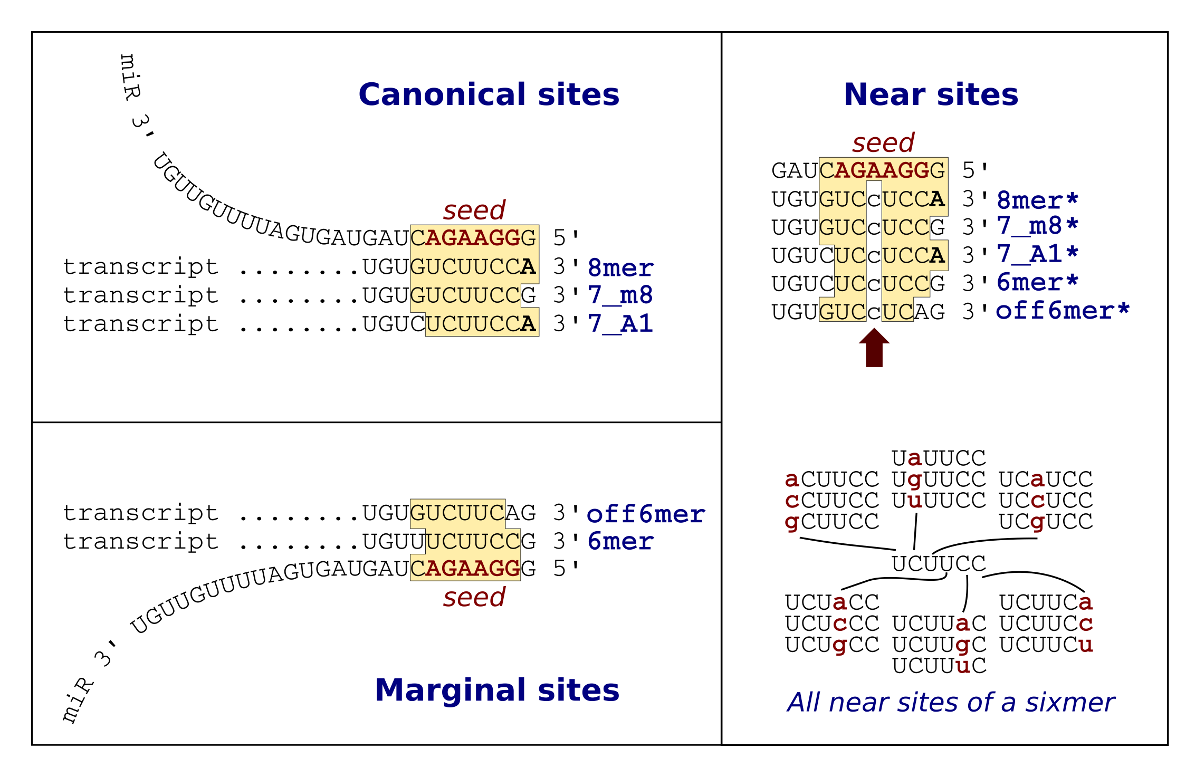
seedVicious is a microRNA target prediction software with several features designed to analyze large datasets, and to study the evolutionary dynamics of target sites. You can predict targets using our web-server. However, it is recommended that you download the stand-alone software to run over larger datasets. You may also explore pre-computed targets for selected species.
Features:
|
Suggestions?Would you like to see a specific feature in future versions of seedVicious? Perhaps you want to predict targets of microRNAs with degenerate positions. Or have precomputed gain/losses of sites for a given set of species? Or you just want to generate regulatory networks based on microRNA/target interactions. Do not hesitate to contact us. There's no such thing as a bad idea. |
| Antonio Marco, 2017, 2018, University of Essex, amarco.bio@gmail.com - University of Essex Accessibility Statement |